With Kenneth Ludwig’s Isoplot becoming more and more difficult to use in modern Excel (let alone on a Mac), there is increasing interest in alternatives. One alternative is Pieter Vermeesch’s IsoplotR. In this note, we will have a look at one way you can use IsoplotR from directly in iolite 4.
Requirements
IsoplotR, as the name implies, reimplements much of the Isoplot functionality (and more) in the R programming language. So, the first task is getting R installed. This can be done following the recommendations for your platform and will not be covered in detail here.
The next task is getting IsoplotR installed in this R environment. With a working R installation, this should be as easy as starting the R interpreter and running:
1
2
|
install.packages('IsoplotR')
|
Of course, in iolite, we have an embedded python interpreter, but no R interpreter. So, the next task is installing the rpy2 python module. You can install this package using the procedure described in an earlier note.
If everything goes according to plan, we should now be ready to call IsoplotR functions from iolite’s python interpreter!
Using IsoplotR to calculate a concordia age
As an example, we will go through compiling the required data, using IsoplotR to calculate a concordia age, and then retrieving the output.
The first thing we need to do is compile all of the measurements to feed into IsoplotR. One way we can do that is as follows:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
|
import pandas as pd
# Choose a group
group = data.selectionGroup('Z_Plesovice')
# Compile the measurements into separate lists
r68 = [data.result(s, data.timeSeries('Final Pb206/U238')).value() for s in group.selections()]
r68s = [data.result(s, data.timeSeries('Final Pb206/U238')).uncertaintyAs2SE() for s in group.selections()]
r75 = [data.result(s, data.timeSeries('Final Pb207/U235')).value() for s in group.selections()]
r75s = [data.result(s, data.timeSeries('Final Pb207/U235')).uncertaintyAs2SE() for s in group.selections()]
rho = [data.associatedResult(s, 'rho 206Pb/238U v 207Pb/235U').value() for s in group.selections()]
# Now create a pandas data frame from those lists
df = pd.DataFrame({
'x': r75,
'sx': r75s,
'y': r68,
'sy': r68s,
'rho': rho
})
|
Now we have all of our results for the ‘Z_Plesovice’ selection group in a pandas data frame called ‘df’. The next step is to convert this into something R, and more importantly, IsoplotR can use. To do that, we’ll make use of some functionality in rpy2 and IsoplotR.
1
2
3
4
5
6
7
8
9
10
11
12
13
|
from rpy2 import robjects as ro
from rpy2.robjects.packages import importr
from rpy2.robjects import pandas2ri
from rpy2.robjects.conversion import localconverter
ir = importr('IsoplotR')
# Use rpy2's functionality to convert a pandas data frame to an R data frame
with localconverter(ro.default_converter + pandas2ri.converter):
rdf = ro.conversion.py2ri(df)
# Now use IsoplotR's functionality to convert that data frame into the format it wants
ird = ir.read_data_data_frame(rdf)
|
We are most of the way there at this stage! Now we just need to call a function in IsoplotR and get the output. We can do that as follows:
1
2
|
out = ir.concordia_age(ird)
age = out[3][0]
|
It is as easy as that. There are many different things output by the concordia age calculation, but the indices used (3 and 0) will be the age in Ma.
Using IsoplotR to plot the concordia age
We can also make one small change to instead plot the concordia diagram:
1
2
|
ir.concordia(ird, show_age=1)
|
This will open a window with a plot showing the measurements as well as the concordia age.
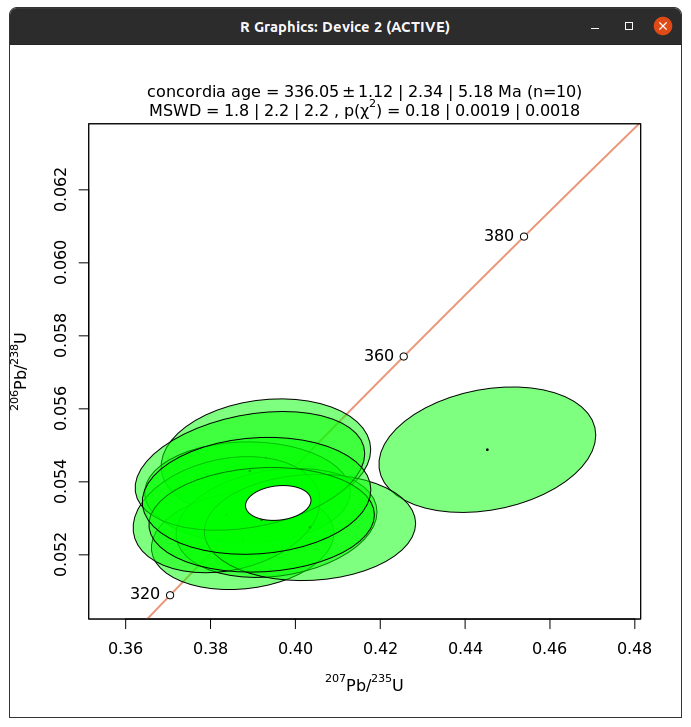
This works fine, but please note that there may be some problems with the resulting window. At least in testing this on Linux, the window cannot be closed.
Putting it all together
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
|
from rpy2 import robjects as ro
from rpy2.robjects.packages import importr
from rpy2.robjects import pandas2ri
from rpy2.robjects.conversion import localconverter
import pandas as pd
ir = importr('IsoplotR')
group = data.selectionGroup('Z_Plesovice')
r68 = [data.result(s, data.timeSeries('Final Pb206/U238')).value() for s in group.selections()]
r68s = [data.result(s, data.timeSeries('Final Pb206/U238')).uncertaintyAs2SE() for s in group.selections()]
r75 = [data.result(s, data.timeSeries('Final Pb207/U235')).value() for s in group.selections()]
r75s = [data.result(s, data.timeSeries('Final Pb207/U235')).uncertaintyAs2SE() for s in group.selections()]
rho = [data.associatedResult(s, 'rho 206Pb/238U v 207Pb/235U').value() for s in group.selections()]
df = pd.DataFrame({
'x': r75,
'sx': r75s,
'y': r68,
'sy': r68s,
'rho': rho
})
with localconverter(ro.default_converter + pandas2ri.converter):
rdf = ro.conversion.py2ri(df)
ird = ir.read_data_data_frame(rdf)
y = ir.concordia_age(ird)
print('Concordia age = %f'%(y[3][0]))
# Uncomment below to display concordia diagram
#ir.concordia(ird, show_age=1)
|
Click here to discuss.
