The 4.4.x series of iolite updates started with a doozy – our new local database plugin – and continued with a myriad of smaller features and bug fixes. Read on to find out more!
Highlights
Database
Our relatively new database plugin allows you to collate results and metadata from iolite (3 and 4) sessions, as well as from Excel, into a sqlite database. This database is wrapped in a simple user interface that allows you to query the results using all the power of SQL and create reports or plots with the output. Since it is just a sqlite file, it can even be used in a variety of 3rd party software.
Here is a simple example, just importing 1 session, doing a simple query to select the Plesovice analyses and then plotting a concordia age.
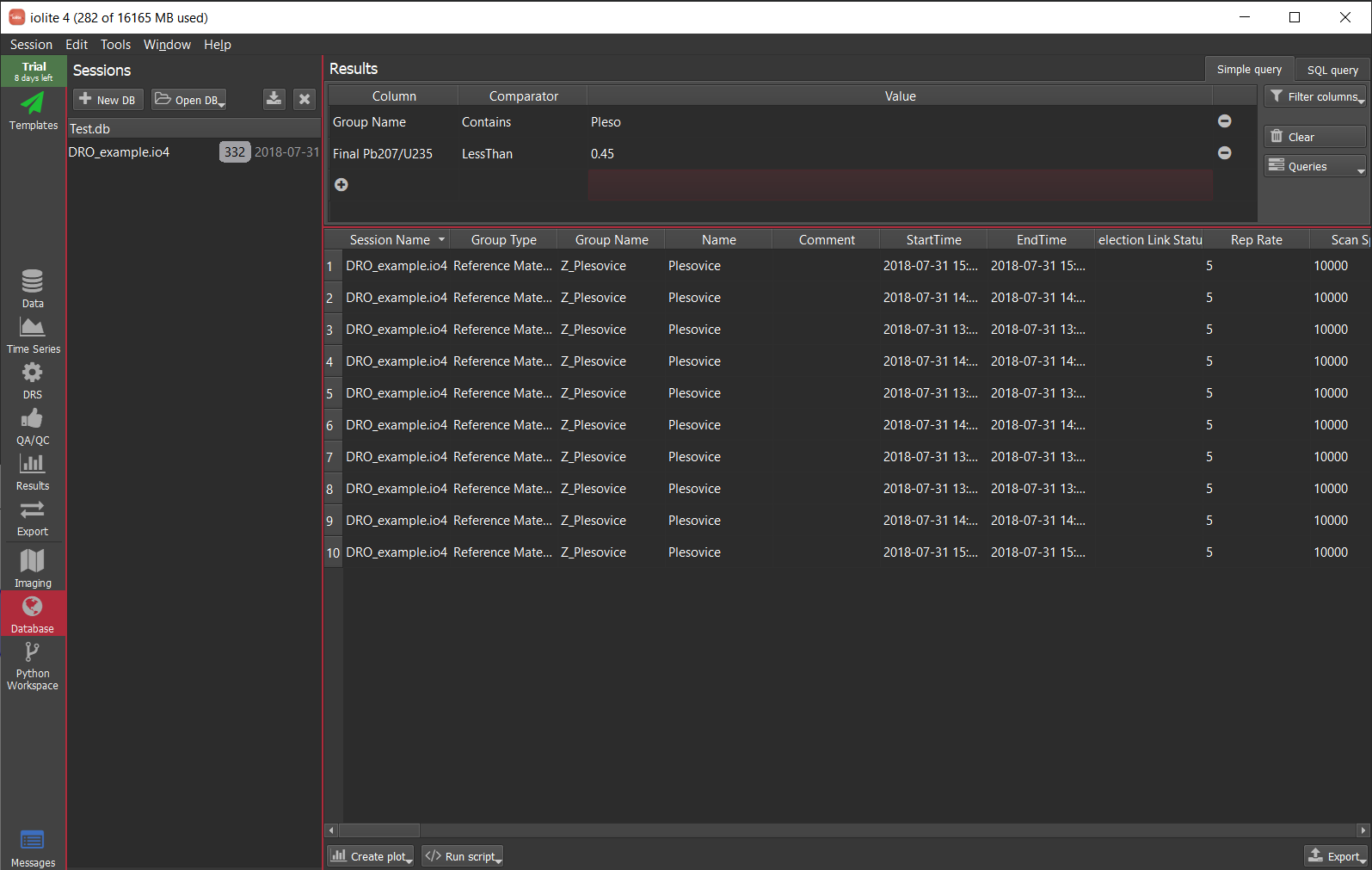
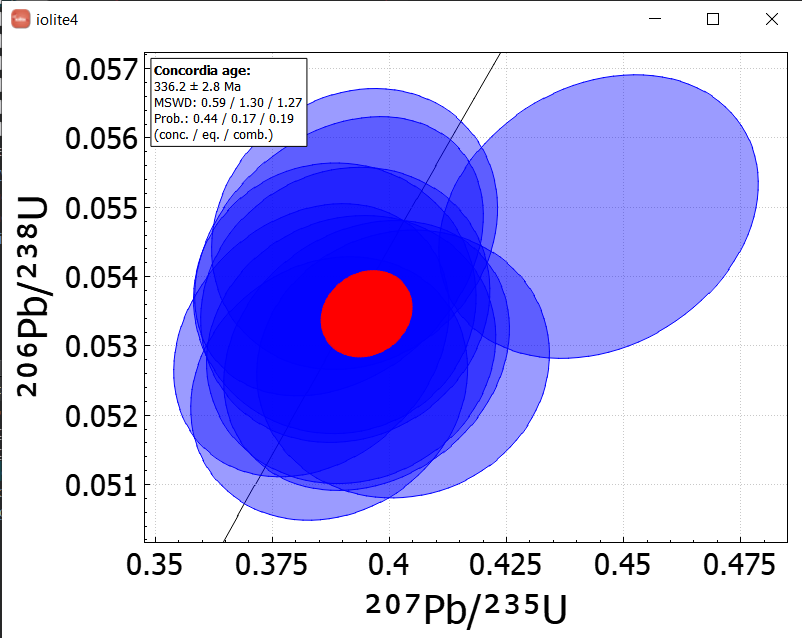
The example below is using much more data - 2121 analyses from 44 sessions collected over nearly 3 years - to look at long term trends in reference materials. We imagine using a database in this way will help our users detect analytical issues and properly assess long term uncertainties, among other things.
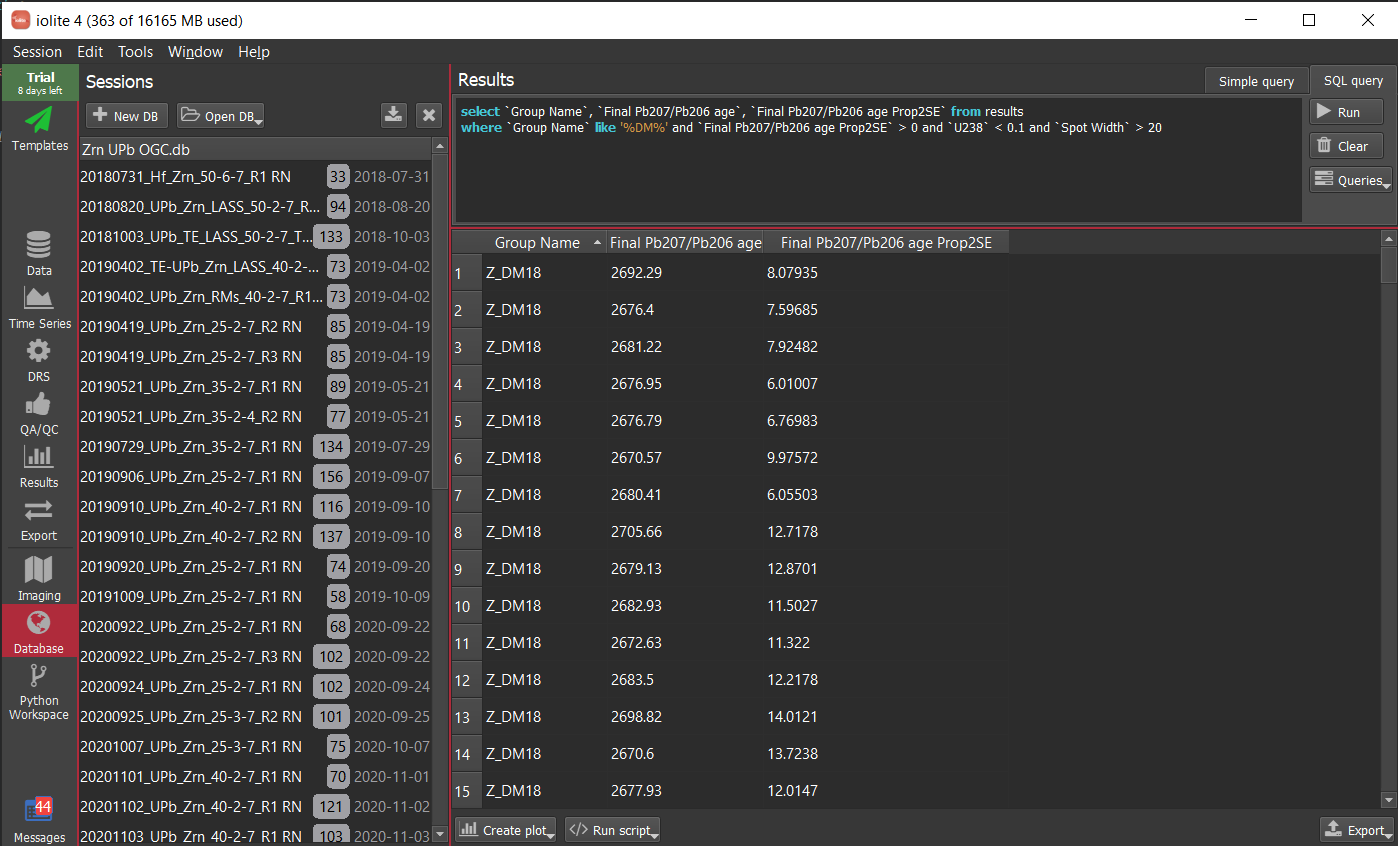
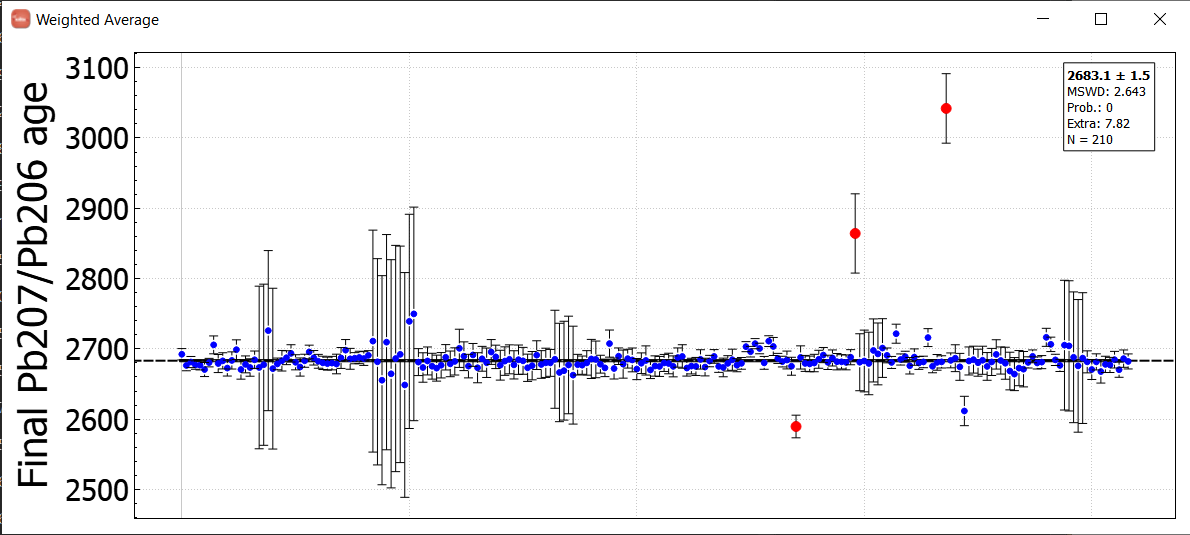
For more information on how this feature works and what it can do, see the documentation or the recent webinar.
Reference material data in session files
A common problem we have in support is that users have gone and created a variety of custom reference material (RM) files and then they send us their session file to investigate some problem. Since we don’t have the RM files we either need to then ask for all of the RM .json files or fill in the missing data from GeoReM. We can do better!
Starting with v4.4.8, when a session is saved, all of the local RM data is stored in the session file. When loading a session, if the RM data in the session file does not match what iolite knows about, it will ask if you want to import the session RM data. This solves our problem in support, but also makes it much easier for users to share session files without worrying about RM data differences.
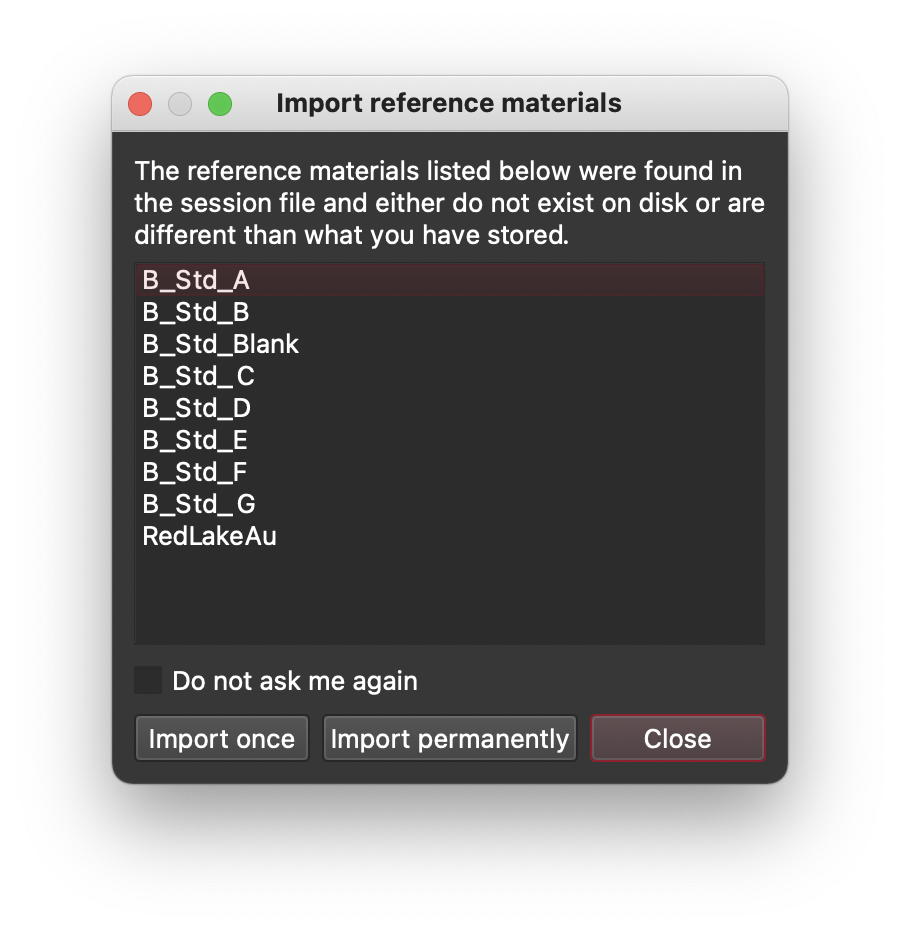
Note that we offer the ability to import the session RM data one time only (i.e. it is loaded in memory but not stored on disk in iolite’s RM folder) or forever (i.e. it is copied to disk, potentially overwriting existing data).
Expanded python api
A variety of features were added or improved in the python api during the v4.4.x series of updates. Some of my favorites include:
- Quickly reading HDF5 data using
data.getH5Array(...)ordata.getH5Attr(...). - Getting time series data by mass using
data.timeSeriesByMass(...). - Importing IS values using
data.importISValues(...). - Save functionality in IolitePlotPyInterface, e.g.
plot.savePdf(...). - Exposed ChannelsComboBox, e.g.
from iolite.ui import ChannelsComboBox. - Exposed OverlayMessage, e.g.
from iolite.ui import OverlayMessage. - Adding/removing IolitePloyPyInterface graphs to/from the legend, e.g.
graph.removeFromLegend(). - Adding color scales to IolitePloyPyInterface, e.g.
plot.addColorScale(...). - And last but not least, creating 3d plots, e.g.:
|
|
which produces the following plot:
Release notes
New features and enhancements:
- Introducing iolite’s new Database feature! It collates your iolite experiments to create a database of results. You can filter, sort and visualize your long term results, as well as keep an eye on your reproducibility and inter-session statistics. There is some introductory documentation available here
- Added a background to the color scale in the imaging detailed 2d view for better readability
- Added an option to hide measurements below LOD in the QA/QC secondary check
- Added condensation temperatures to the elements database
- Small changes to discordia age QA/QC module to expand the ranges plotted
- Modified discordia intercept age finding code to be more similar to Isoplot and be more robust
- Changed the opacity of ellipses plotted in the XY results view
- Changed image matrix export so its orientation matches on screen
- Fixed up scale bar drawing for imaging
- Added a way to restore defaults to REEView for element and normalization configurations
- Expanded python API
- You can now load internal standard (IS) values for reference materials (where the IS value varies) in processing templates
- You can now jump to a channel’s properties from the Time Series View by right-clicking its name
- Added the ability to import one database into another
- Added tooltips to database query menus to show the full query
- Changed the way the time range is zoomed when clicking on a selection in the time series view list – now zooming to the original extent of the selection if created from samples
- Added support for editing certain columns in a database
- Improvements to database export from processing templates
- Added an option to not error when create selections processing template action produces no selections
- Added support for iCap mass shifting
- Added an experimental synchrotron data importer
- Added an option to export that allows the error being reported to fallback to internal when propagated is missing
- Added the ability to move selections by duration in a processing template
- Added minimum/maximum durations when creating selections in processing templates
- Added basic undo and redo for adding and deleting actions in processing templates
- Added more command line options, including the ability to run processing templates or python
- Improved presentation of logarithmic color scales
- Some minor additions and improvements to the python API
- Added undo/redo for deleting channels
- Improved icpTOF importer
Recent fixes:
- Fixed some issues with loading HDF5 files on Windows
- Added some improvements for Trace Elements Next DRS
- Provided some minor improvements to imaging
- Database: now when importing the same session file (with or without possible changes), iolite will now just warn you that you are importing duplicates of the same selections rather than reporting an error
- Fixed an issue during export that affected column headers
- Fixed some issues with re-syncing laser log files
- Added some improvements for handling channel names in split stream experiments
- Provided some minor improvements to imaging
- Fixed an issue with instrument names in split stream experiments that might lead to data not being loaded
- Database: Quick fix for plotting against start and end times
- Fixed issue with the zip file importer that didn’t allow file to be selected in Data Browser
- You can now add configurations to the REEView
- Fixed error that would be reported during Processing Template import action if a csv file could not be imported. Now just reports a warning and continues
- Improved laser log sync algorithm in Processing Templates to now use the same as that in the Data Browser
- Fixed issue with masking in Baseline Subtract DRS
- Fixed a bug in automatic selections from channels where a match on the first point could cause an spurious interval
- Fixed an issue where channels were not cleared before running DRS in processing template
- Fixed masking in Hf and UComPbine data reduction schemes
- Some fixes for imaging related to pixel sizes and region pixmaps
- Fixed possible exception in imaging when a channel previously imaged is no longer available
- Fixed unnecessary import in database concordia script
- Fixed VizualAge problem preventing 204 correction if 208 and 232 were not measured
- Fixed a possible issue with imaging ROI subdivide by probability
- Fixed an issue with imaging data table export not honouring SD vs SE setting
- Some minor fixes associated with databases
- Fixed an issue when importing IS values where incorrect matches might occur
- Fixed an issue with custom plot settings save and restore
- Fixed an issue with processing template export configurations
- Fixed U-Pb ppm channel calculations for channels named by mass rather than element
- Fixed an issue preventing some DRS settings from being editable in processing templates
- Fixed an issue with processing template import file selector only allowing .csv
- Fixed an issue with processing template save session action
- Fixed an issue with the database data provider used for plotting
- Fixed possible hang when zooming around dashed lines (e.g. splines in the time series view)
- Fixed an issue in U-Pb for split stream data where Pb/Th was measured on both instruments
- Fixed an issue causing crashing in the channels menu (used in imaging among other places) that only seemed to affect Apple silicon Macs
- Fixed an issue with the laser log combiner when entries had the same time stamp
- Fixed a potential issue setting trace element IS data with out selecting the entire row
- Fixed a potential issue with processing template import log action not prompting for the file when the method was set to ‘Prompt user’
- Fixed a potential issue with starting new trials
Click here to discuss.